Managing Dependencies
If you are in a hurry:
- Want to use your own modules and packages in a flow? See Structuring Projects.
- Want to use a Python library in a flow? See Managing Libraries.
- Want to use
uvwith Metaflow? See usinguv - Want to use or build a specific Docker image? See Defining Custom Containers.
New in Metaflow 2.15.8: You can use uv to manage dependencies.
A Metaflow flow is defined in a single Python file. Besides the flow code
itself, a typical flow has many software dependencies, that is, other
modules,
packages,
and 3rd party libraries like pandas or pytorch of which the flow is composed.
While the question of dependency management may seem rather mundane, it is a critically important part of production-quality projects for a number of reasons:
Flows need a stable execution environment. You don't want to get unexpected results because a package abruptly changed under the hood. Each project may have its own set of dependencies that need to be isolated from others.
It should be possible to make flows reproducible. Anyone should be able to retrieve a flow, rerun it, and get similar results. To make this possible, a flow can't depend on libraries that are only available on your laptop.
Remote execution of tasks requires that all dependencies can be reinstantated on the fly in a remote environment. Again, this is not possible if the flow depends on libraries that are only available on your laptop.
Metaflow includes robust but flexible machinery for managing dependencies. You can get started without hassle and harden the project gradually to provide it with a stable, reproducible execution environments.
Behind the scenes, Metaflow does much more than just pip install'ing packages
on the fly. If you are curious to learn what and why,
see Packaging Internals and the @pypi
announcement blog post.
Unpacking a Metaflow project
Let's consider the layers that make up a Metaflow project:
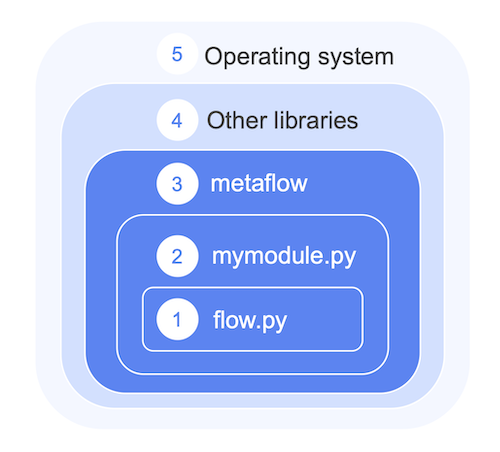
A flow file, e.g.
flow.pydefines the flow DAG. The file may contain arbitrary user-defined code, making it easy to get started just with a single file.As the project grows, it is convenient to structure the project as multiple modules and packages, instead of including hundreds of lines in a single file. Metaflow packages local Python dependencies like this automatically.
Crucially, Metaflow packages Metaflow itself for remote execution so that you don't have to install it manually when using
@batchand@kubernetes. Also, this guarantees that all tasks use the same version of Metaflow consistently.External libraries can be included via the
@pypiand@condadecorators. Behind the scenes, these decorators take care of dependency resolution, virtual environment creation, and package snapshotting automatically.Ultimately, all the code executes on top of an operating system which defines low-level drivers and security functionality. If needed, much of this can be configured via a custom Docker image.
Note that you can get started with local development without having to worry about any of this. Metaflow uses libraries you have installed locally like any other Python project. Dependency management becomes relevant immediately when you want to start scaling out to the cloud or you want to deploy the flow in production.
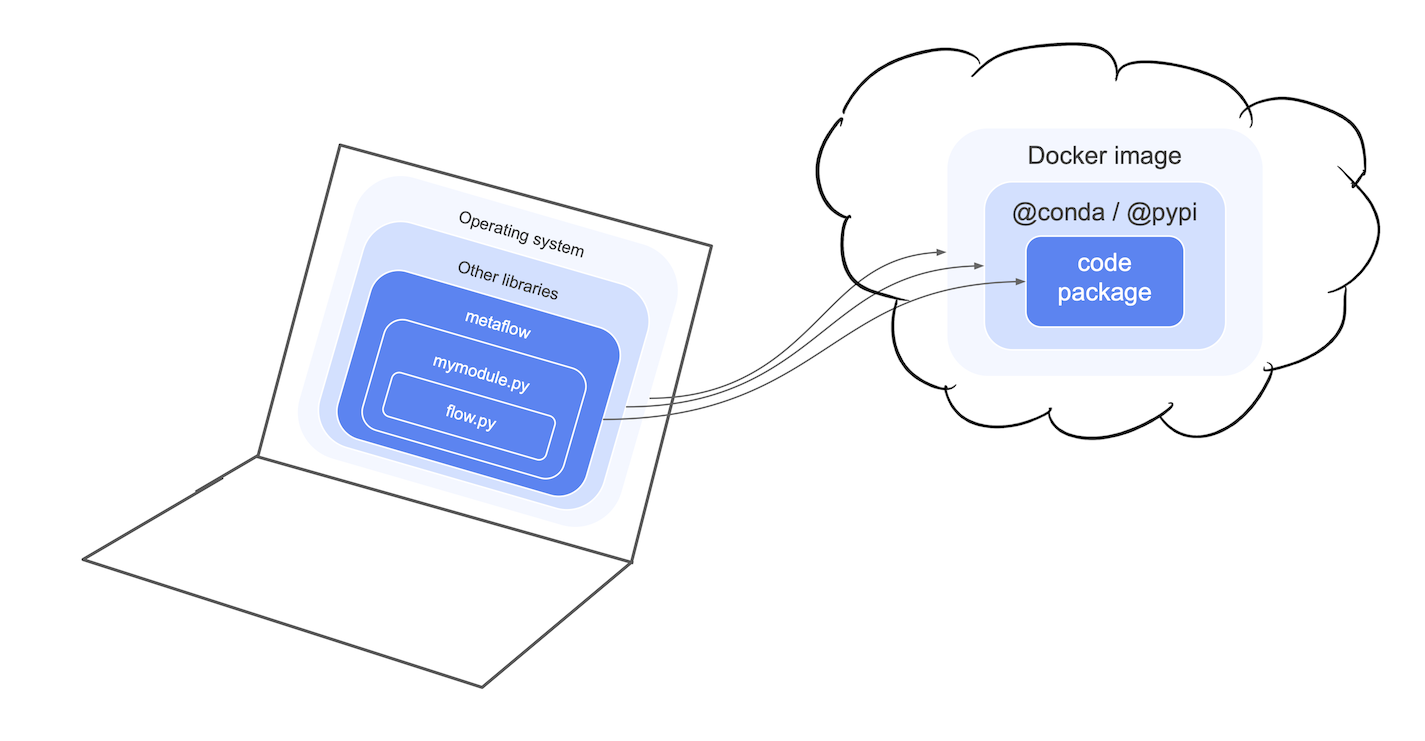
Packaging projects for remote execution
To execute tasks outside your local environment, Metaflow snapshots the layers 1-3(dark blue) automatically and stores them in a code package. You can inspect the package through the Client API to see the exact code that was executed by a run.
If you are curious, you can inspect the files that would be included in a code package by executing
python myflow.py package list
By the default, the package only includes local Python files, not libraries you
have installed e.g. with pip install manually. To include external libraries,
you need to include them either in a custom Docker
image, specify them in a @pypi or @conda
decorator, or use uv.
This figure illustrates the idea:
Which approach to use?
Let's say your team has a common module special_module.py that is used by
many flows. You could include it in any of the three layers. You could
- Include it next to
flow.pyas a local dependency which Metaflow packages automatically. - Publish it as a Python package and include it with
@pypi. - Include it in a custom Docker image.
Which approach to choose? There may be internal reasons to choose one approach over the others but if you are unusure, here's an easy rule of thumb: Prefer local files for any dependencies that need to change quickly.
If you need to change special_module.py multiple times a day, the overhead of
having to build new packages or Docker files for every change can be
overwhelming. In contrast, if the file rarely changes, all the approaches work
equally well.
The next page focuses on working with local files for rapid development.