Organizing Results
A boring, under-appreciated part of high-quality science (or any project work in general), is keeping results organized. This is the key to effective collaboration, versioning of parallel lines of work, and reproducibility.
The good news is that Metaflow does 80% of this work for you without you having to do anything. This document explains how Metaflow keeps things organized with a concept called namespaces and how you can optionally make results even neater with tags.
Namespaces
As explained in Basics of Metaflow, Metaflow persists all runs and all the
data artifacts they produce. Every run gets a unique run ID, e.g. HelloFlow/546, which
can be used to refer to a specific set of results. You can access these results with the
Client API.
Many users can use Metaflow concurrently. Imagine that Anne and Will are collaborating
on a project that consists of two flows, PredictionFlow and FeatureFlow. As they,
amongst other people, run their versions independently they end up with the following
runs:
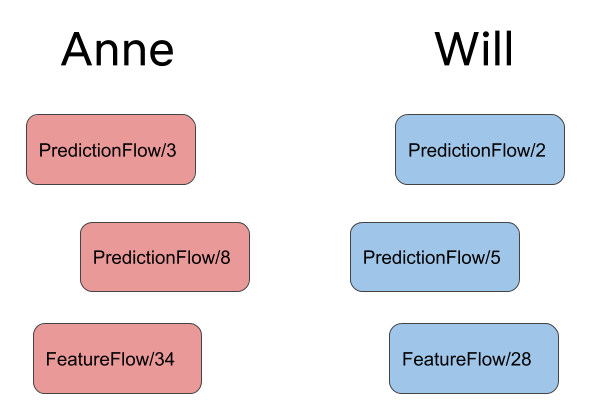
Anne could analyze her latest PredictionFlow results in a notebook by remembering that
her latest run is PredictionFlow/8. Fortunately, Metaflow makes this even easier
thanks to namespaces:

When Anne runs PredictionFlow, her runs are automatically tagged with her
username, prefixed with user:. By default, when Anne uses the Client API,
the API only returns results that are tagged with user:anne. Instead of having to
remember the exact ID of her latest run, she can simply say:
library(metaflow)
run <- flow_client$new('PredictionFlow')$latest_run
For Anne, this will return 'PredictionFlow/8'. For Will, this will return
'PredictionFlow/5'.
Switching Namespaces
Namespaces are not about security or access control. They help you to keep results organized. During development, organizing results by the user who produced them is a sensible default.
You can freely explore results produced by other people. In a notebook (for example), Anne can write
library(metaflow)
set_namespace('user:will')
run <- flow_client$new('PredictionFlow')$latest_run
to see Will's latest results, in this case, 'PredictionFlow/5'.
You can also access a specific run given its ID directly:
library(metaflow)
run <- flow_client$new('PredictionFlow')$run("5")
However, this will fail for Anne, since PredictionFlow/5 is in Will's namespace. An
important feature of namespaces is to make sure that you can't accidentally use someone
else's results, which could lead to hard-to-debug, incorrect analyses.
If Anne wants to access Will's results, she must do so explicitly by switching to Will's namespace:
library(metaflow)
set_namespace('user:will')
run <- flow_client$new('PredictionFlow')$run("5")
In other words, you can use the Client API freely without having to worry about using incorrect results by accident.
If you use the Client API in your flows to access results of other flows, you can use
the --namespace flag on the command line to switch between namespaces. This is a
better approach than hardcoding a set_namespace() function call in the code that
defines your Metaflow workflow.
Global Namespace
What if you know a run ID, but you don't know whose namespace it belongs to? No worries, you can access all results in the Metaflow universe in the global namespace:
library(metaflow)
set_namespace(NULL)
run <- flow_client$new('PredictionFlow')$run("5")
Running set_namespace(NULL) allows you to access all results without limitations. Be
careful though: relative references like latest_run make little sense in the global
namespace since anyone can produce a new run at any time.
Resuming across namespaces
The resume command is smart enough to work
across production and personal namespaces. You can resume a production workflow
without having to do anything special with namespaces.
You can resume runs of other users, and you can resume any production runs. The results of your resumed runs are always created in your personal namespace.
Production Namespaces
During development, namespacing by the username feels natural. However, when you schedule your flow to run automatically, runs are not related to a specific user anymore. It is typical for multiple people to collaborate on a project that has a canonical production version. It is not obvious which user "owns" the production version.
Moreover, it is critical that you, and all other people, can keep experimenting on the
project without having to worry about breaking the production version. If the production
flow ran in the namespace of any individual, relative references like latest_run could
break the production easily as the user keeps executing experimental runs.
As a solution, by default the production namespace is made separate from the usernamespace:
-9e825aaf633bd9c3557c35b34291c647.png)
Isolated production namespaces have three main benefits:
- Production tokens allow all users of Metaflow to experiment freely with any project without having to worry about accidentally breaking a production deployment. Even if they ran step-functions create, they could not overwrite a production version without explicit consent, via a shared production token, by the person who did the previously authorized deployment.
- An isolated production namespace makes it easy to keep production results separate from any experimental runs of the same project running concurrently. You can rest assured that when you switch to a production namespace, you will see only results related to production - nothing more, nothing less.
- By having control over the production namespace, you can alter data that is seen by production flows. For instance, if you have separate training and prediction flows in production, the prediction flow can access the previously built model as long as one exists in the same namespace. By changing the production namespace, you can make sure that a new deployment isn't tainted by old results.
If you are a single developer working on a new project, you don't have to do anything
special to deal with production namespaces. You can rely on the default behavior of
step-functions create.
Production tokens
When you deploy a Flow to production for the first time, Metaflow creates a new, isolated production namespace for your production flow. This namespace is identified by a production token, which is a random identifier that identifies a production deployment, e.g. production:PredictionFlow3 above. You can examine production results in a notebook by switching to the production namespace.
If another person wants to deploy a new version of the flow to production, they must use the same production token. You, or whoever has the token, are responsible for sharing it with users who are authorized to deploy new versions to production. This manual step should prevent random users from deploying versions to production inadvertently.
After you have shared the production token with another person, they can deploy a new version with
- Bash
- RStudio
Rscript production_flow.R step-functions create --authorize TOKEN_YOU_SHARED_WITH_THEM
...
step(step = "end",
...)
%>%
run(step_fucntions = "create",
authorize = "TOKEN_YOU_SHARED_WITH_THEM")
They need to use the --authorize option only once. Metaflow stores the token for them
after the first deployment, so they need to do this only once.
Resetting a production namespace
If you call step-functions create again, it will deploy an updated version of your
code in the existing production namespace of the flow.
Sometimes the code has changed so drastically that you want to recreate a fresh namespace for its results. You can do this as follows:
- Bash
- RStudio
Rscript production_flow.R step-functions create --generate-new-token
...
step(step = "end",
...)
%>%
run(step_fucntions = "create",
generate_new_token = TRUE)
This will deploy a new version in production using a fresh, empty namespace
Tagging
The user: tag is assigned by Metaflow automatically. In addition to automatically
assigned tags, you can add and remove arbitrary tags in objects. Tags are an excellent
way to add extra annotations to runs, tasks etc., which makes it easier for you and
other people to find and retrieve results of interest.
An easy way to add tags is the --tag command line option. You can add multiple tags
with multiple --tag options. For instance, this will annotate a HelloFlow run with a
tag crazy_test.
- Terminal
- RStudio
Rscript helloworld.R run --tag crazy_test
# Replace run() in helloworld.R with
# run(tag = c("crazy_test"))
# and execute in RStudio
The --tag option assigns the specified tag to all objects produced by the run: the run
itself, its steps, tasks, and data artifacts.
Accessing Tags
You can access runs (or steps or tasks) with a certain tag easily using the Client API:
library(metaflow)
run <- flow_client$new("HelloFlow").runs_with_tags("crazy_test")[[1]]
This will return the latest run of HelloFlow with a tag crazy_test in your
namespace. Filtering is performed both based on the current set_namespace() and the
tag filter.
You can also filter by multiple tags:
library(metaflow)
run <- flow_client$new("HelloFlow").runs_with_tags("crazy_test", "date:2020-06-01")[[1]]
This requires that all the tags listed, and the current namespace, are present in the object.
You can see the set of tags assigned to an object with the .tags property. In the
above case, run.tags would return a set with a string crazy_test amongst other
automatically assigned tags.
Tags as Namespaces
Let's consider again the earlier example with Anne and Will. They are working on their
own versions of PredictionFlow but they want to collaborate on FeatureFlow. They
could add a descriptive tag, say xyz_features, to FeatureFlow runs.
Now, they can easily get the latest results of FeatureFlow regardless of the user who
ran the flow:
library(metaflow)
set_namespace('xyz_features')
run <- flow_client$new('FeatureFlow')$latest_run
This will return FeatureFlow/34 which happened to be run by Anne. If Will runs the
flow again, his results will be the latest results in this namespace.
We encourage you to use a combination of namespaces, domain-specific tags, and filtering by tags to design a workflow that works well for your project.
Accessing Current IDs in a Flow
Tagging and namespaces, together with the Client API, are the main ways for accessing results of past runs. Metaflow uses these mechanisms to organize and isolate results automatically, so in most cases you don't have to do anything.
However, in some cases you may need to deal with IDs explicitly. For instance, if your flow interacts with external systems, it is a good idea to inform the external system about the identity of the run, so you can trace back any issues to a specific run. Also IDs can come in handy if you need to version externally stored data.
For this purpose, Metaflow provides a singleton object current that represents the
identity of the currently running task. Use it in your FlowSpec to retrieve current
IDs of interest:
library(metaflow)
start <- function(self){
print(paste0("flow name: ", current("flow_name")))
print(paste0("run id: ", current("run_id")))
print(paste0("origin run id: ", current("origin_run_id")))
print(paste0("step name: ", current("step_name")))
print(paste0("task id: ", current("task_id")))
print(paste0("pathspec: ", current("pathspec")))
print(paste0("username: ", current("username")))
}
metaflow("CurrentFlow") %>%
step(step="start",
r_function=start,
next_step="end") %>%
step(step="end") %>%
run()
You can see the output
2020-06-19 21:19:03.387 [198/start/1139 (pid 64853)] [1] "flow name: CurrentFlow"
2020-06-19 21:19:03.387 [198/start/1139 (pid 64853)] [1] "run id: 198"
2020-06-19 21:19:03.387 [198/start/1139 (pid 64853)] [1] "origin run id: "
2020-06-19 21:19:03.388 [198/start/1139 (pid 64853)] [1] "step name: start"
2020-06-19 21:19:03.388 [198/start/1139 (pid 64853)] [1] "task id: 1139"
2020-06-19 21:19:03.389 [198/start/1139 (pid 64853)] [1] "pathspec: CurrentFlow/198/start/1139"
2020-06-19 21:19:03.389 [198/start/1139 (pid 64853)] [1] "username: jge"
2020-06-19 21:19:08.400 [198/start/1139 (pid 64853)] Task finished successfully.
The current singleton also provides programmatic access to the CLI option
--origin-run-id used by the resume within
your flow code.
For regular run invocations, the value of current("origin_run_id") is NULL.
If a user explicitly overrides the CLI option --origin-run-id, the current singleton
would reflect that value. Suppose we invoked resume for the above script to re-run
everything from start without explicitly overriding the CLI option origin-run-id, we
can see the value chosen by Metaflow using the current singleton:
- Bash
- RStudio
Rscript current_flow.R resume start
# Replace run() in current_flow.R with
# run(resume = "start")
# and execute in RStudio
You should see the origin_run_id used by the resume in the output (the exact value
for you might be different):
"origin run id: 198"